Even faster Wild Cluster Bootstrap Inference in R via WildBootTests.jl 🚀
In the last few months, I have spent quite a bit of time working with a Julia package - WildBootTests.jl - and towards integrating it into fwildclusterboot via the awesome JuliaConnectoR.
Now fwildclusterboot 0.8 has made its way to CRAN, so I thought it would be time to convince you to install Julia and to run your wild bootstraps through WildBootTests.jl - but of course, still from R. 😄
‘WildBootTests.jl’
A few months ago, I wrote a blog post on the outstanding speed gains that can be achieved by the ‘fast’ wild cluster bootstrap algorithm, which is implemented in R in the fwildclusterboot package. In some benchmarks, fwildclusterboot's boottest() function ran the wild cluster bootstrap more than 1000 times faster than sandwich::vcovBS! Amazingly, it turns out that WildBootTests.jl is even faster than the algorithm in fwildclusterboot or Stata’s boottest module.
But we have all heard that Julia is fast, so it’s maybe no too surprising that WildBootTests.jl shines with speed. In my opinion, there are at least three other reason why it’s worth to try out WildBootTests.jl:
WildBootTests.jlis very memory efficientWildBootTests.jlimplements the wild cluster bootstrap for IV regression by Davidson & MacKinnon, and it’s blazing fast.WildBootTests.jlallows for wild cluster bootstrapped F-tests 1
Before I’ll start showcasing WildBootTests.jl, I will quickly describe how to access WildBootTests.jl from R via fwildclusterboot.
Getting Started
To run WildBootTests.jl through fwildclusterboot, both Julia and WildBootTests.jl need to be installed. The best place to install Julia is its homepage - just follow the instructions you find there. To facilitate getting started with everything else, I have written a small package, JuliaConnectoR.utils, which aims to help to install WildBootTests.jl and all its dependencies from within R and to connect Julia and R via the JuliaConnectoR package.
So after installing Julia, simply run 2
devtools::install_github("s3alfisc/JuliaConnectoR.utils")
library(JuliaConnectoR.utils)
# connect julia and R
connect_julia_r()
# install WildBootTests.jl
install_julia_packages("WildBootTests.jl")and you’re good to go and can wild cluster bootstrap via WildBootTests.jl.
A first bootstrap via WildBootTests.jl
The only required things left are a) a regression model to bootstrap and b) to specify boottest()'s boot_algo function argument, so let’s start with simulating some data and fitting a regression:
library(fwildclusterboot)
library(modelsummary)
N <- 100000
data <- fwildclusterboot:::create_data(
N = N,
N_G1 = 50, icc1 = 0.1,
N_G2 = 20, icc2 = 0.8,
numb_fe1 = 10,
numb_fe2 = 10,
seed = 123,
weights = 1:N
)
lm_fit <- lm(
proposition_vote ~ treatment + as.factor(Q1_immigration) + as.factor(Q2_defense),
data
)By default, boottest() will run fwildclusterboot's R-implementation of the wild cluster bootstrap. To run through WildBootTests.jl, you have to specify the boot_algo function argument:
boot_r <-
fwildclusterboot::boottest(
lm_fit,
clustid = c("group_id1"),
B = 9999,
param = "treatment",
seed = 3,
nthreads = 1
)
boot_julia <-
fwildclusterboot::boottest(
lm_fit,
clustid = c("group_id1"),
B = 9999,
param = "treatment",
seed = 3,
nthreads = 1,
boot_algo = "WildBootTests.jl"
)Note that the first call of WildBootTests.jl takes around 10-20 seconds to, which is due to the fact that the Julia code being run is initially pre(-JIT)-compiled.
Of course, it is good to see that the R implementation of the fast wild cluster bootstrap and WildBootTests.jl lead to (almost) identical inferences:
msummary(
list(
"R" = boot_r,
"Julia" = boot_julia
),
estimate = "{estimate} ({p.value})",
statistic = "[{conf.low}, {conf.high}]"
)| R | Julia | |
|---|---|---|
| 1*treatment = 0 | 0.003 (0.002) | 0.003 (0.001) |
| [0.001, 0.005] | [0.001, 0.006] | |
| Num.Obs. | 100000 | 100000 |
| R2 | 0.159 | 0.159 |
| R2 Adj. | 0.159 | 0.159 |
| AIC | -46670.1 | -46670.1 |
| BIC | -46470.4 | -46470.4 |
| Log.Lik. | 23356.067 | 23356.067 |
Now that I have shown how it works, let’s proceed to the next question: why should you install Julia and run WildBootTests.jl if the ‘fast’ algorithm is already implemented in ‘native’ R?
Reason I: It is extremely fast
In short: fwildclusterboot's R algo is fast, but we all know that Julia is faster, and WildBootTests.jl turns the speed of the wild cluster bootstrap from 10 to 11:

Turning the overdrive from 10 to 11 with WildBootTests.jl
In a blog post from a few months ago, I claimed that fwildclusterboot was 1000 times faster than sandwich's vcovBS() function. Below, I run the same problem, once for boot_algo = "R" and once with boot_algo = "WildBootTests.jl":
B <- 9999
bench <- bench::mark(
boot_r =
fwildclusterboot::boottest(
lm_fit,
clustid = c("group_id1"),
B = B,
param = "treatment",
seed = 3,
nthreads = 1
),
boot_julia =
fwildclusterboot::boottest(
lm_fit,
clustid = c("group_id1"),
B = B,
param = "treatment",
seed = 3,
nthreads = 1,
boot_algo = "WildBootTests.jl"
),
iterations = 3,
check = FALSE
)
bench## # A tibble: 2 x 6
## expression min median `itr/sec` mem_alloc `gc/sec`
## <bch:expr> <bch:tm> <bch:tm> <dbl> <bch:byt> <dbl>
## 1 boot_r 737ms 758ms 1.32 304MB 6.60
## 2 boot_julia 266ms 342ms 2.85 97MB 4.75For a problem that took boottest()'s R algorithm around 1.5 seconds, WildBootTests.jl finishes in around half a second. WildBootTests.jl is around 3 times faster than fwildclusterboot's R implementation. Benchmarked against sandwich::vcovBS, WildBootTests.jl is around 4500x faster!
In general, WildBootTests.jl is - after compilation - at least an order of a magnitude faster than fwildclusterboot's ‘R’ algorithm. Below are benchmarks from a regression with \(N=10000\) observations and \(k=20\) covariates.
While the R algorithm is competitive for ‘small’ problems, with a growing number of clusters and observations problems, WildBootTests.jl clearly outperforms. For the most complex problem with \(G = 1000\) clusters and \(B =99999\) iterations, WildBootTests.jl finishes in around 8 seconds, while the “R-algo” on 4 cores needs around twice as long. On one core, it runs for more than 40 seconds. And note that WildBootTests.jl can also be run in parallel - check out JuliaConnectoR.utils::set_julia_nthreads() for instructions on how to set the number of threads for Julia from within R.
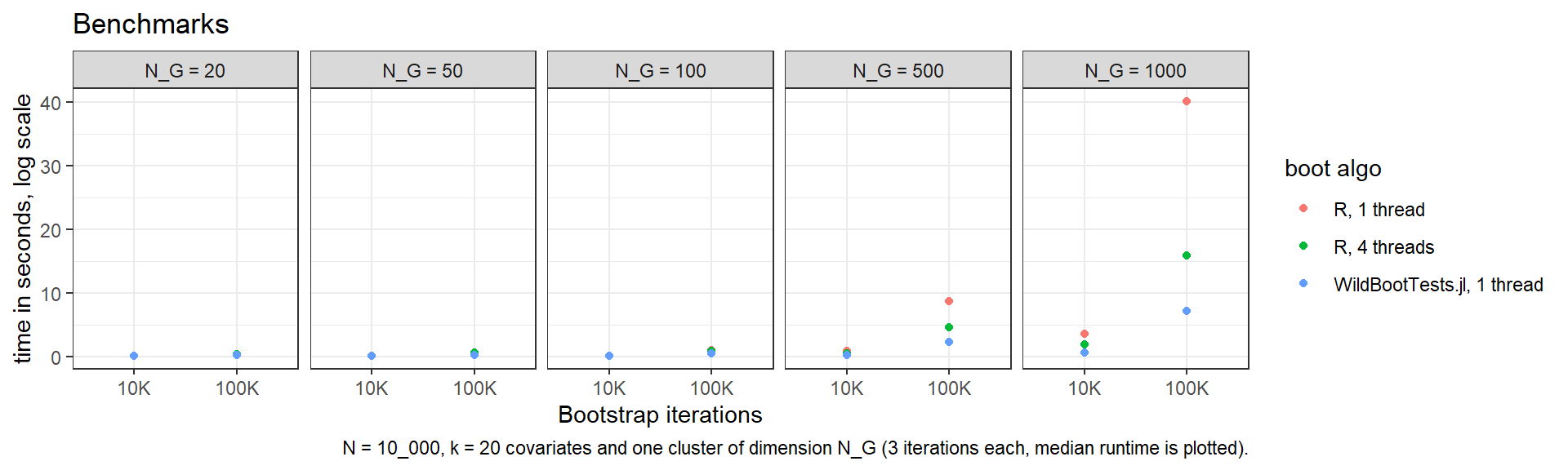
WildBootTests.jl is very memory-efficient
A second selling point of WildBootTests.jl is that it is less memory-demanding. As fwildclusterboot's R algorithm is fully vectorized, a large bootstrap weights matrix \(v^{*}\) of dimensions \(G \times B\) is created and stored at once. As programming in Julia is much more encouraging towards writing loops, the large matrix does not need to be created at once, which prohibits the occasional out-of-memory error that the R algorithm encounters when \(G\) and \(B\) get too large.
WildBootTests.jl implements the Wild Bootstrap for IV
Third (and maybe most importantly), WildBootTests.jl offers additional functionality that has previously not been available to R users - most notably, it implements the WRE bootstrap for instrumental variable estimation from Davidson & MacKinnon. David Roodman, who is WildBootTests.jl's author, has spend a lot of effort on optimizing the WRE’s performance, and as a result, it is blazing fast.
nlsw88 <- haven::read_dta("http://www.stata-press.com/data/r8/nlsw88.dta")
head(nlsw88)## # A tibble: 6 x 17
## idcode age race married never_married grade collgrad south smsa c_city
## <dbl> <dbl> <dbl+l> <dbl+l> <dbl> <dbl> <dbl+lb> <dbl> <dbl+l> <dbl>
## 1 1 37 2 [bla~ 0 [sin~ 0 12 0 [not ~ 0 1 [SMS~ 0
## 2 2 37 2 [bla~ 0 [sin~ 0 12 0 [not ~ 0 1 [SMS~ 1
## 3 3 42 2 [bla~ 0 [sin~ 1 12 0 [not ~ 0 1 [SMS~ 1
## 4 4 43 1 [whi~ 1 [mar~ 0 17 1 [coll~ 0 1 [SMS~ 0
## 5 6 42 1 [whi~ 1 [mar~ 0 12 0 [not ~ 0 1 [SMS~ 0
## 6 7 39 1 [whi~ 1 [mar~ 0 12 0 [not ~ 0 1 [SMS~ 0
## # ... with 7 more variables: industry <dbl+lbl>, occupation <dbl+lbl>,
## # union <dbl+lbl>, wage <dbl>, hours <dbl>, ttl_exp <dbl>, tenure <dbl>Currently, fwildclusterboot allows to run the WRE after IV-estimation via ivreg::ivreg()3
library(ivreg)
# test that coefficient on tenure = 0, clustering errors by industry
iv_fit <- ivreg(wage ~ tenure + ttl_exp + collgrad | union + ttl_exp + collgrad, data = nlsw88)
boot_iv <-
boottest(
iv_fit,
param = "tenure",
B = 9999,
clustid = "industry"
)
summary(iv_fit)##
## Call:
## ivreg(formula = wage ~ tenure + ttl_exp + collgrad | union +
## ttl_exp + collgrad, data = nlsw88)
##
## Residuals:
## Min 1Q Median 3Q Max
## -16.3074 -2.8239 -0.3707 2.3584 37.8481
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 4.9615 0.6206 7.994 2.27e-15 ***
## tenure 0.7409 0.1945 3.809 0.000144 ***
## ttl_exp -0.2323 0.1413 -1.644 0.100439
## collgrad 2.9808 0.2658 11.216 < 2e-16 ***
##
## Diagnostic tests:
## df1 df2 statistic p-value
## Weak instruments 1 1864 29.61 5.97e-08 ***
## Wu-Hausman 1 1863 22.55 2.20e-06 ***
## Sargan 0 NA NA NA
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 4.803 on 1864 degrees of freedom
## Multiple R-Squared: -0.3255, Adjusted R-squared: -0.3277
## Wald test: 117.6 on 3 and 1864 DF, p-value: < 2.2e-16summary(boot_iv)## boottest.ivreg(object = iv_fit, clustid = "industry", param = "tenure",
## B = 9999)
##
## Hypothesis: 1*tenure = 0
## Observations: 1855
## Bootstr. Type: rademacher
## Clustering: 1-way
## Confidence Sets: 95%
## Number of Clusters: 12
## ## term estimate statistic p.value conf.low conf.high
## 1 1*tenure = 0 0.741 2.491 0.022 0.202 1.798Additionally, WildBootTests.jl supports wild cluster bootstrapping of multiple joint hypotheses. E.g. to jointly test the null that tenure = 0 and collgrad = 0 after a linear regression model, one would use fwildclusterboot's new mboottest() function:
lm_fit <- lm(wage ~ tenure + ttl_exp + collgrad, data = nlsw88)
boot_q2 <-
mboottest(
lm_fit,
R = clubSandwich::constrain_zero(2:4, coef(lm_fit)),
B = 9999,
clustid = "industry"
)
summary(boot_q2)## mboottest.lm(object = lm_fit, clustid = "industry", B = 9999,
## R = clubSandwich::constrain_zero(2:4, coef(lm_fit)))
##
## Hypothesis: Multivariate mboottest
## Observations: 2217
## Bootstr. Type: rademacher
## Clustering: 1-way
## Number of Clusters: 12
## ## teststat p_val
## 1 56.79246 0.015625Conclusion
WildBootTests.jl is outstanding software - do check it out!
P.S. If you want all you wild bootstraps to run through WildBootTests.jl, you can simply set a global variable at the top of your script:
setBoottest_boot_algo("WildBootTests.jl")Afterwards, each call to boottest() will simply default to boot_algo = "WildBootTests.jl" unless explicitly stated otherwise.
implemented in R via the wildmeta package.↩︎
of course you also have to install
fwildclusterboot, which you can install from CRAN, github, and r-universe.↩︎I plan to add support for IV estimation via
fixestandlfein the next weeks / months.↩︎